Principle Component Analysis was performed using the Iris dataset.
# Load data
data(iris)
head(iris, 3)## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa# log transform
log.ir <- log(iris[, 1:4])
ir.species <- iris[, 5]
# apply PCA - scale. = TRUE is highly
# advisable, but default is FALSE.
ir.pca <- prcomp(log.ir,
center = TRUE,
scale. = TRUE)# print method
print(ir.pca)## Standard deviations:
## [1] 1.7124583 0.9523797 0.3647029 0.1656840
##
## Rotation:
## PC1 PC2 PC3 PC4
## Sepal.Length 0.5038236 -0.45499872 0.7088547 0.19147575
## Sepal.Width -0.3023682 -0.88914419 -0.3311628 -0.09125405
## Petal.Length 0.5767881 -0.03378802 -0.2192793 -0.78618732
## Petal.Width 0.5674952 -0.03545628 -0.5829003 0.58044745# plot method
plot(ir.pca, type = "l")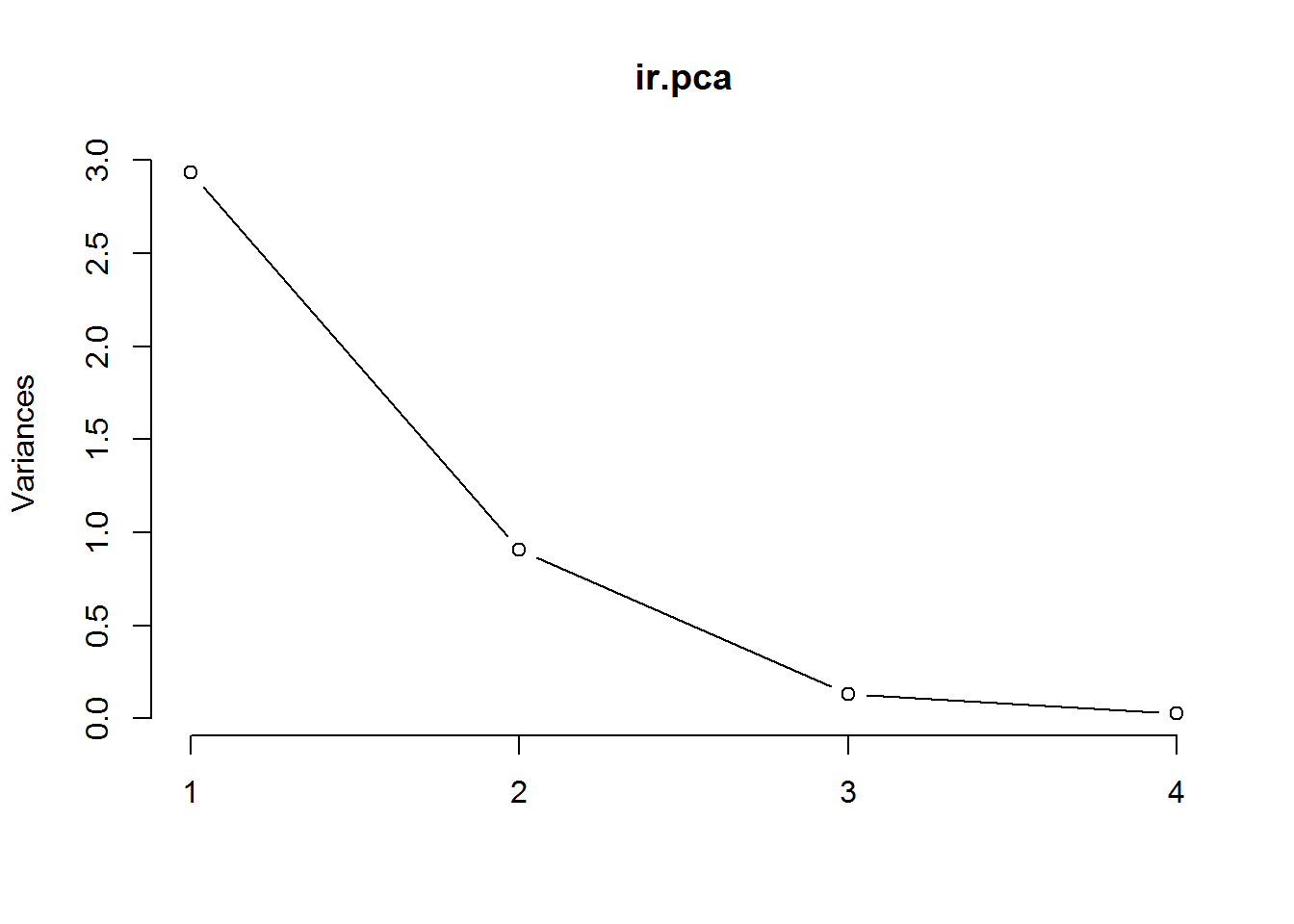
# summary method
summary(ir.pca)## Importance of components:
## PC1 PC2 PC3 PC4
## Standard deviation 1.7125 0.9524 0.36470 0.16568
## Proportion of Variance 0.7331 0.2268 0.03325 0.00686
## Cumulative Proportion 0.7331 0.9599 0.99314 1.00000# Predict PCs
predict(ir.pca,
newdata=tail(log.ir, 2))## PC1 PC2 PC3 PC4
## 149 1.0809930 -1.01155751 -0.7082289 -0.06811063
## 150 0.9712116 -0.06158655 -0.5008674 -0.12411524#if (!require(devtools)) install.packages("devtools")
if (!require(ggbiplot)) install_github("ggbiplot", "vqv")## Loading required package: ggbiplot## Loading required package: ggplot2## Warning: package 'ggplot2' was built under R version 3.3.3## Loading required package: plyr## Loading required package: scales## Loading required package: gridlibrary(devtools)## Warning: package 'devtools' was built under R version 3.3.3library(ggbiplot)
g <- ggbiplot(ir.pca, obs.scale = 1, var.scale = 1,
groups = ir.species, ellipse = TRUE,
circle = TRUE)
g <- g + scale_color_discrete(name = '')
g <- g + theme(legend.direction = 'horizontal',
legend.position = 'top')
print(g)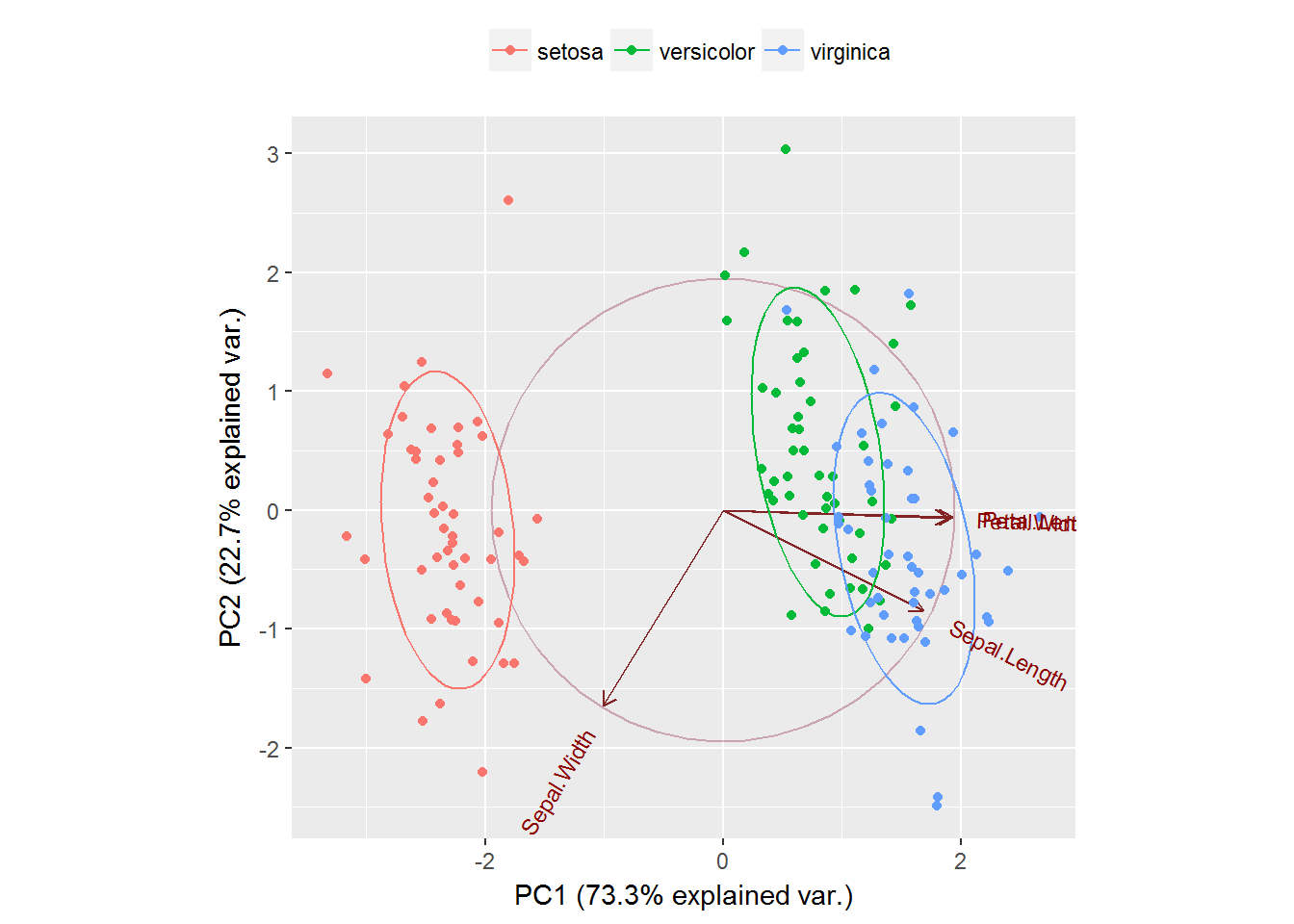
Reference: https://www.r-bloggers.com/computing-and-visualizing-pca-in-r/

Share this post
Twitter
Google+
Facebook
Reddit
LinkedIn
StumbleUpon
Email